Basic R Programming
R vs RStudio (1)

is a language and environment for statistical computing and graphics.
Provides a wide variety of statistical (linear and nonlinear modelling, classical statistical tests, time-series analysis, classification, clustering, etc.) and publication-quality graphical techniques. And many more!
FREE (under GNU-GPL license).

Rstudio is an integrated development environment (IDE) of R
Provides extensible environments for compiling other languages (e.g. Python, Shell, LaTeX, etc.) and engines (e.g. knitr, Jupyter, quarto, etc.)
FREE.
R vs RStudio (2)
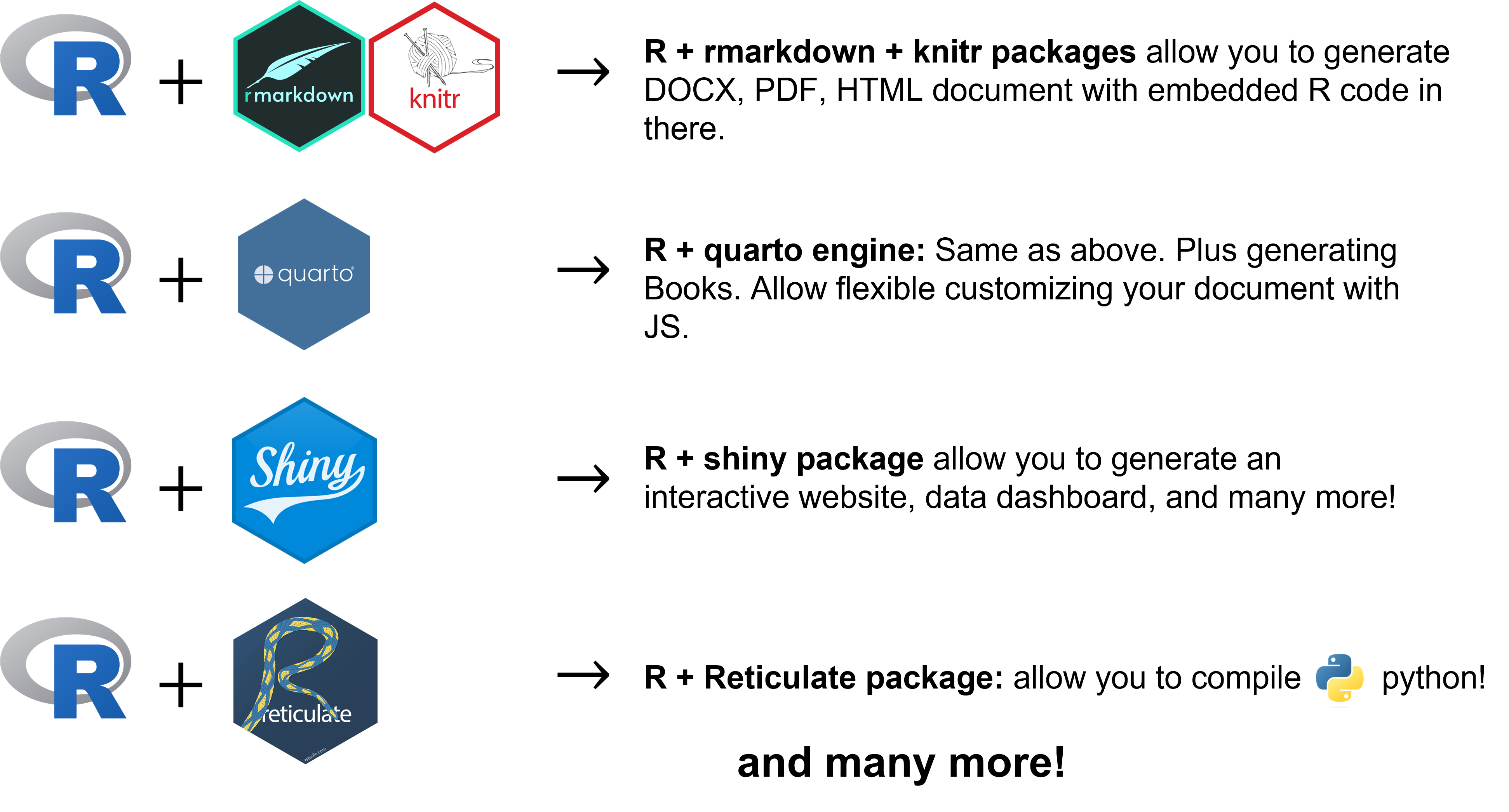
R: versatility
Loading R libraries (1)
Common way to load library
Loading R libraries (2)
More efficient way to load multiple libraries at once with pacman:
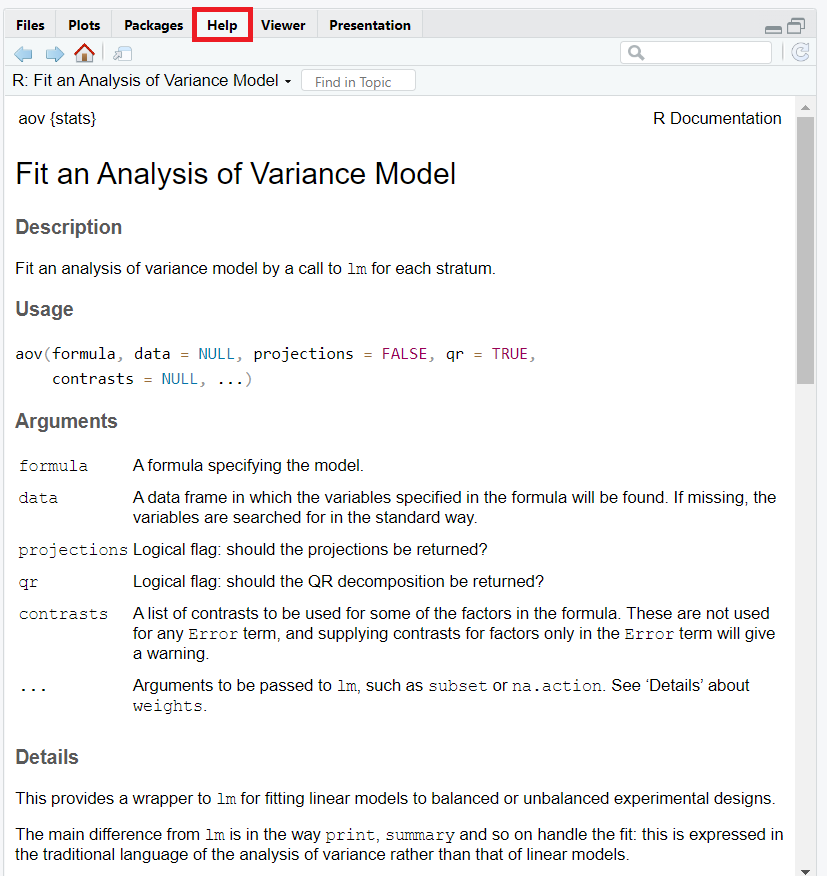
Accessing Help & Documentation in RStudio
RStudio provides built-in documentation of all functions you have installed from libraries. For example, you would like to access documentation page of function aov, simply type in the console as follows:
You can access this documentation from the Help pane. The documentation includes an explanation of the arguments, background theorem, and references for the function aov.
Tip
There are 2 recommended repositories allow you to access all documentation online, RDocumentation.org and rdrr.io. These repositories contain all the documentation for all the functions available in R, even if you have never installed it!
Data types
R has 5 data types:
- Character
- Numeric
- Integer
Data structure (1)
Vectors
is a row of strings (can be numbers, characters, logicals , or mix of it), and also known as a 1-dimensional array. R uses function
cto declare vectors:
Matrices
is a 2-dimensional array, we use the function
matrixto declare matrix in R as follow.[,1] [,2] [1,] 9 4 [2,] 2 5 [3,] 3 6[1] 4[,1] [,2] [1,] 90 40 [2,] 20 50 [3,] 30 60[,1] [,2] [1,] 9 4 [2,] 2 5 [3,] 20 6
Data structures (2)
Data frames
A data frame is a matrix in which rows and columns are named. A data frame is more flexible and compatible for further data manipulation and export as a spreadsheet. Also, data frame can be calculated like matrix.
# Create a data frame t <- data.frame( name = c("gene1", "gene2", "gene3", "gene4"), cond_1 = c(20, 18, 0, 0), cond_2 = c(1, 2, 100, 120) ) # Access element in data frame t[4, 3][1] 120[1] 4 3'data.frame': 4 obs. of 3 variables: $ name : chr "gene1" "gene2" "gene3" "gene4" $ cond_1: num 20 18 0 0 $ cond_2: num 1 2 100 120
Lists
List is a complex object that can store all data types and structures, even list within list!
$one [1] 1 $two [1] 1 2 $five [1] 0.00 0.25 0.50 0.75 1.00$five [1] 0.00 0.25 0.50 0.75 1.00[1] 0.25[1] 0.0 2.5 5.0 7.5 10.0
Data frames (1)
Data frame is a key data structure in R and statistics.
Each row represents observation (genes, protein, taxon, name)
Each column represents variable (measures, treatments, characteristics) of the observation
Each value in a cell represents each data point.
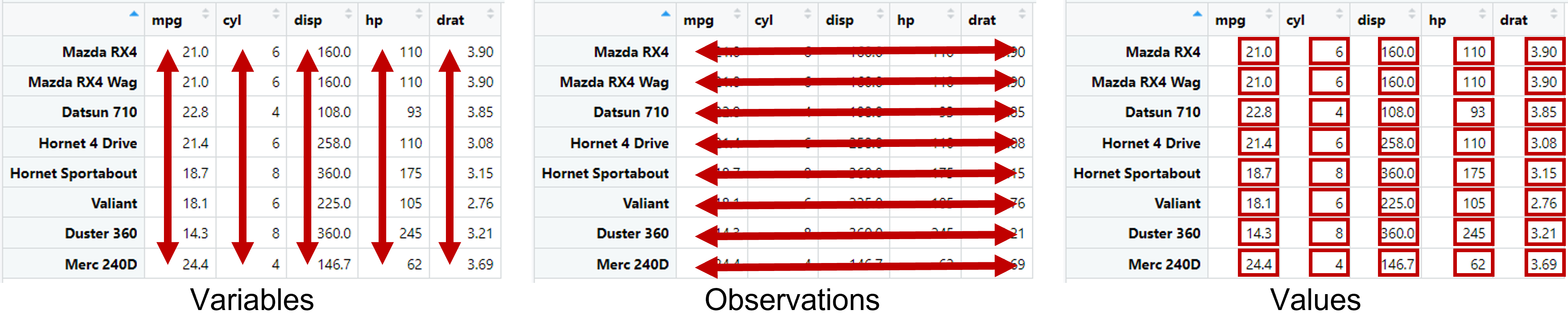
Structure of data frame. Redraw from R for Data Science 2nd edition (Hadley Wickham & Garrett Grolemund).
Data frames (2)
We’ll show structure of the data frames in 2 formats; wide and long formats, using airquality dataset.
Wide format
| Ozone | Solar.R | Wind | Temp | Month | Day |
|---|---|---|---|---|---|
| 41 | 190 | 7.4 | 67 | 5 | 1 |
| 36 | 118 | 8.0 | 72 | 5 | 2 |
| 12 | 149 | 12.6 | 74 | 5 | 3 |
| 18 | 313 | 11.5 | 62 | 5 | 4 |
| NA | NA | 14.3 | 56 | 5 | 5 |
| 28 | NA | 14.9 | 66 | 5 | 6 |
Human-readable data frame
Elegance
Easy to see all values in each observation
One observation is one row
May incompatible for some plots in ggplot2
Long format
| Month | Day | name | value |
|---|---|---|---|
| 5 | 1 | Ozone | 41.0 |
| 5 | 1 | Solar.R | 190.0 |
| 5 | 1 | Wind | 7.4 |
| 5 | 1 | Temp | 67.0 |
| 5 | 2 | Ozone | 36.0 |
| 5 | 2 | Solar.R | 118.0 |
Machine-readable data frame
Simple
Each observation can be more than one row
Compatible to include with metadata table (if any)
ggplot2 ❤️long-format data frame
Managing data frames with dplyr
We can handle data frames with base R, but when you are working with a large data set, speed matters. The dplyr package provides a “grammar” (especially verbs) for data manipulation and for editing data frames.
Frequently used dplyr verbs:
glimpse: skim structure of the data, see every columns in a data frame.select: return a subset of the columns of a data frame, using a flexible notation.filter: extract a subset of rows from a data frame based on logical conditions.arrange: reorder rows of a data frame.rename: rename variables in a data frame.mutate: add new variables/columns or transform existing variables.summarise/summarize: generate summary statistics of different variables in the data frame, possibly within strata.%>%: the “pipe” operator, is used to connect multiple verb actions together into a pipeline.
Common dplyr Function Properties
The first argument must be a data frame to process.
The subsequent arguments describe what to do with the data frame specified in the first argument, and you can refer to columns in the data frame directly without using the
$operator (just use the column names).The return result of a function is a new data frame
For example:
# Load dplyr library
library(dplyr)
# Load airquality dataset
dt <- datasets::airquality
dt_filtered <- filter(dt, Solar.R > 300)
# Show how the data looks like
head(dt_filtered) Ozone Solar.R Wind Temp Month Day
1 18 313 11.5 62 5 4
2 14 334 11.5 64 5 16
3 34 307 12.0 66 5 17
4 30 322 11.5 68 5 19
5 11 320 16.6 73 5 22
6 39 323 11.5 87 6 10dplyr::glimpse
- Create new variable
dt_iristhat stored the data set “iris”.
- Glimpse the data set in
dt_iris

Three species of iris flowers in this data set: setosa, versicolor, and virginica. Figure from https://www.embedded-robotics.com/iris-dataset-classification.
dplyr::select
- We already have data frame
dt_irisfrom earlier practice. Now we will select columns nameSpecies, andPetal.Widthfromdtand store in new variable:dt_sel
- The glimpse the result.
dplyr::filter
filter() is used to subset a data frame, retaining all rows that satisfy your conditions.
From the data set iris stored in data frame dt_iris,
- Now we will filter Species ‘versicolor’.
- From
dt_versicolor, filter the flowers that theSepal.Lengthlonger than or equal to 6
- Then glimpse the result
dplyr::arrange
arrange() orders the rows of a data frame by the values of selected columns.
- In our filtered data frame
dt_vsc_filt, sort theSepal.Lengthcolumn.
- Then, sort the
Petal.Lengthdescendingly.
- Glimpse the result
dplyr::rename
rename() changes the names of individual variables using new_name = old_name syntax.
- From sorted and filtered data frame
dt_vsc_filt_srt, we will rename 2 columns, fromSepal.LengthandPetal.Length, toSLandPL, respectively. Then save to the new data framedt_vsc_renamed.
dplyr::mutate
mutate() creates new columns that are functions of existing variables, as well as modify and delete columns.
- From the previous data frame
dt_vsc_renamed, we’ll calculate the difference between sepal lengthSLand petal lengthPLto the new columnLen_Diff. This can be done with themutate()function as follow.
- Then, use function
summary()to see the distribution of the values using the columnLen_Diff.
Expected result:
Min. 1st Qu. Median Mean 3rd Qu. Max.
0.9 1.5 1.9 1.8 2.0 2.3 dplyr::%>%
The pipeline operator %>% (pronounce: pipe) is very handy for bundling dplyr verbs and creating complex syntax for processing data. For example:
- Instead of using
dplyrverbs and storing the new variables line by line, we can bundle them and use%>%. All operations associated with%>%are stored in one variable.
From the syntax above:
We loaded the iris data set to the variable
iris_df.Then rename the column name with
rename()function.Then calculate the difference of sepal length and petal length using
mutate()function.And keep the difference that are greater than 1 using
filter()function.All of these verbs are operated and store in one variable,
iris_df.
dplyr::summarize
summarise() returns one row for each combination of grouping variables. It will contain one column for each grouping variable and one column for each of the summary statistics that you have specified.
- We’ll load original
datasets::iristo the new data framedt2_iris.
- Of these 3 species, calculate mean and standard deviation of petal length using
mean()andsd(), respectively. Then calculate the standard deviation of mean (SEM) of the petal length.
\[ SEM = \frac{SD}{\sqrt{n}} \]
| Species | num_flowers | Mean_PL | SD_PL | SEM_PL |
|---|---|---|---|---|
| setosa | 50 | 1.462 | 0.1736640 | 0.0245598 |
| versicolor | 50 | 4.260 | 0.4699110 | 0.0664554 |
| virginica | 50 | 5.552 | 0.5518947 | 0.0780497 |
Plotting systems in R
Plotting is an important tool for understanding data properties, finding patterns in the data, suggesting modeling strategies for our data, and communicating what we have found in our data. Many plotting systems available in R such as:
Base graphic conventional way, same as implementing graphical visualizations in the S language. You can only draw on the plot, and append another plot to it.
Grid graphic or Grobs (graphical objects), not used to create statistical graphs per se, but are insanely useful in combining and laying out multiple graphic devices.
Lattice Plots uses lattice graphics to implement the Trellis graphics system. Also known as an improved version of Base Plot.
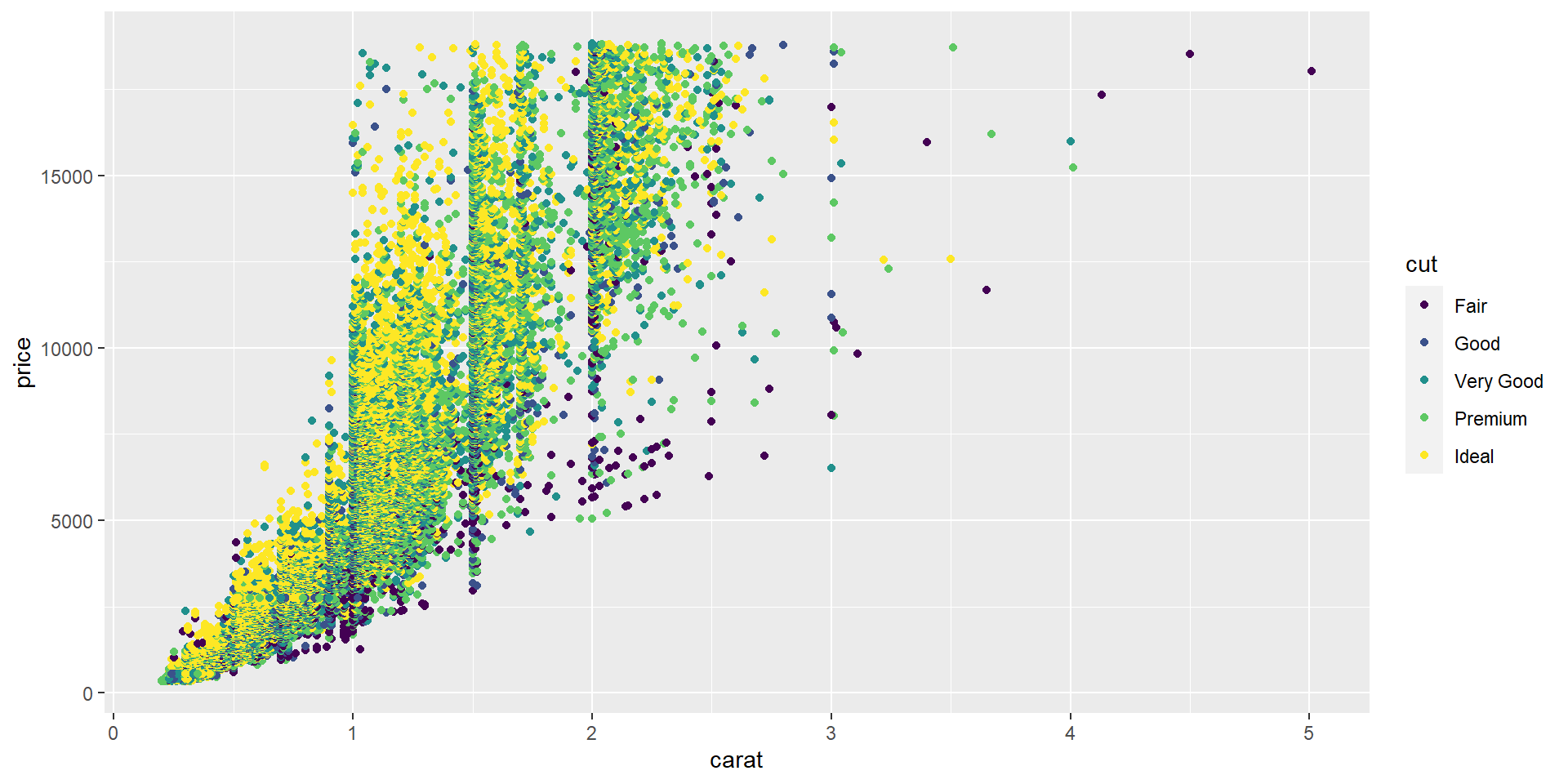
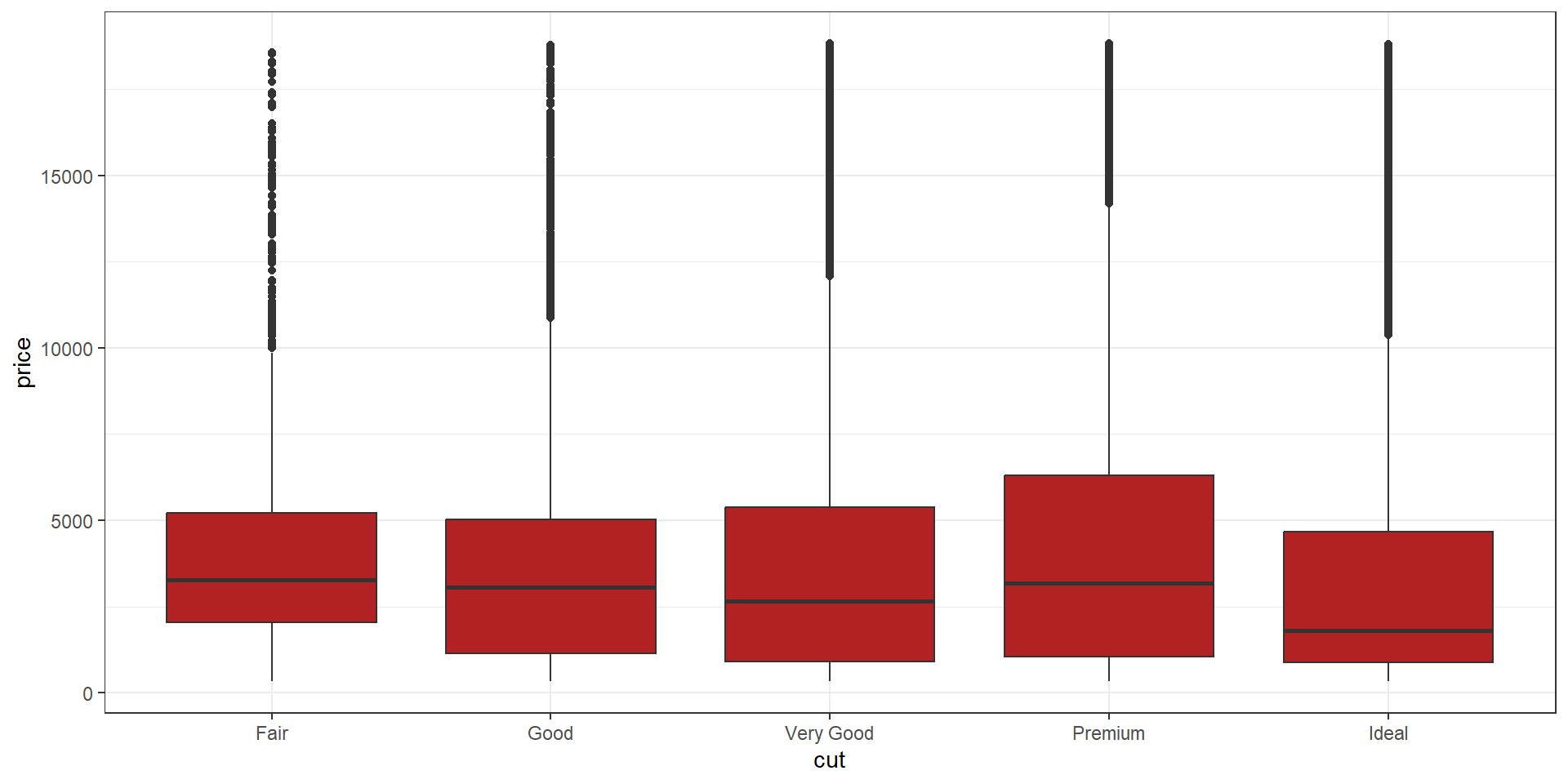
ggplot2 improves base and lattice graphics. The graphics are drawn using grids, which allows you to manipulate their appearance at many levels.
htmlwidgets provides a common framework for accessing web visualization tools from R. Userful for creating interactive plots for publishing on websites.
plotly is a popular javascript visualization toolkit with an R interface. It is a great tool if you want to create interactive graphics for HTML documents or websites.
Another graphic systems, ComplexHeatmap (Gu 2022), will be used in this workshop as well.
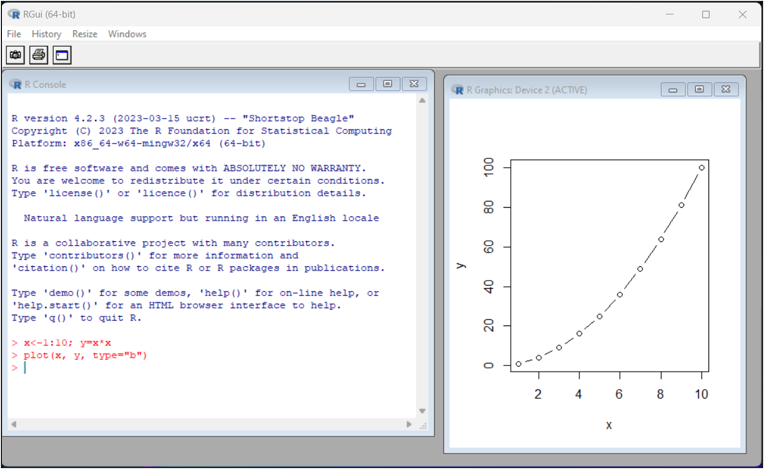
Base graphics (1)
Using library graphics, plain and simple plot functions in R is usually called R base plot. The syntax is shown as follow:
Base graphics (2)
The following lines create a plot from data frame t.
# Creating a data frame
t <- data.frame(x = c(11,12,14),
y = c(19,20,21),
z = c(10,9,7))
# Creating a new plot
plot(t$x, type = "b", ylim = range(t), col = "red")
# Adding new graphic to the plot
lines(t$y, type = "s", col = "blue")
# Adding another graphic to the plot
points(t$z, pch = 20, cex = 2, col = "green")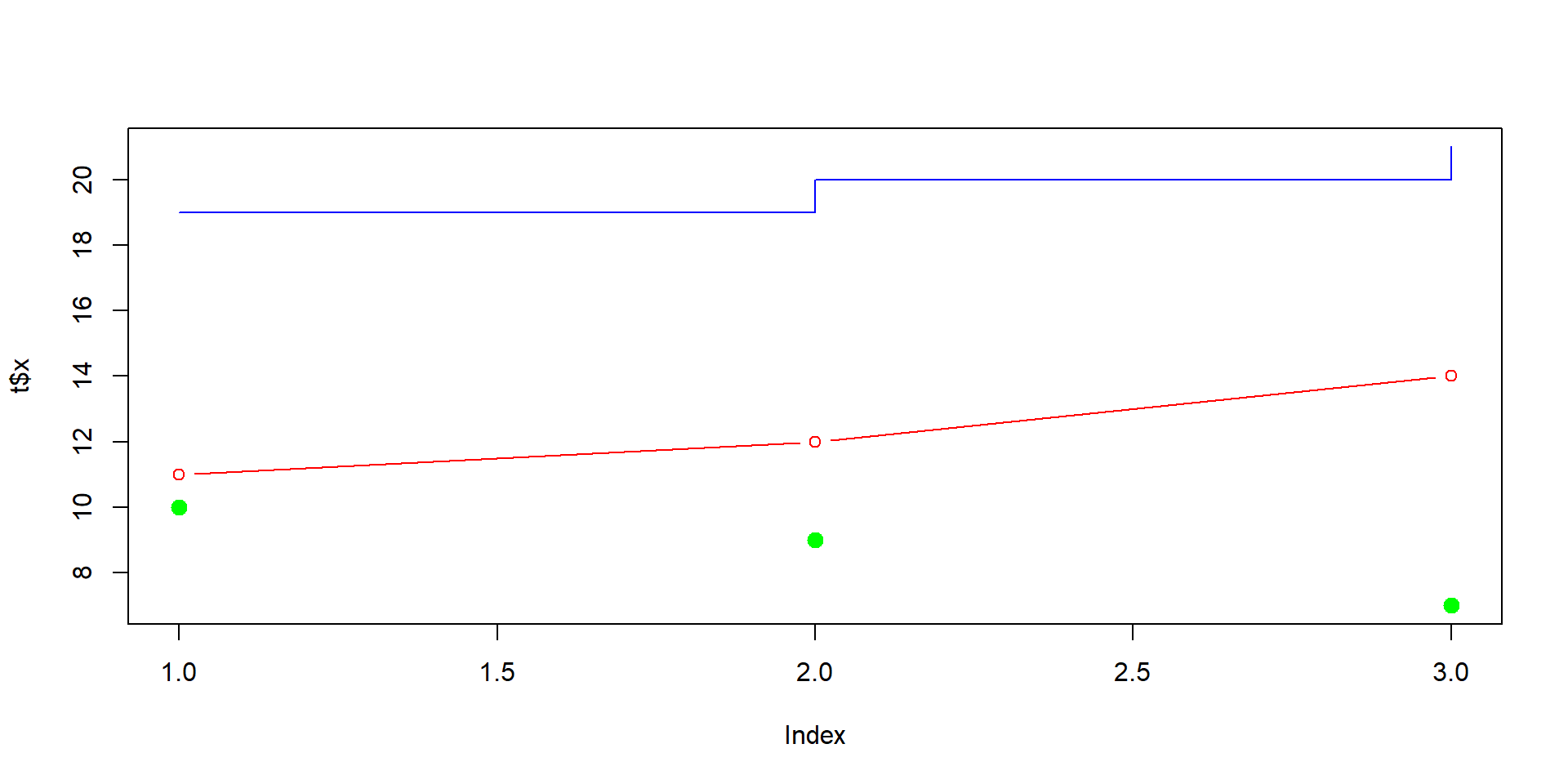
Lattice graphic
Attempts to improve R’s basic graphs by providing better presets and the ability to display multivariate relationships. In particular, the package supports the creation of grid graphs - graphs that show a variable or the relationship between variables as a function of one or more other variables.
ggplot2
The ggplot2 package is an R package for creating graphs or plots of statistical data. With ggplot2, you can compose graphs by combining independent components based on the Grammar of Graphics.
We’ll mainly use ggplot2 and other graphic libraries in this workshop 🙂
Tidyverse
References
- A (very) short introduction to R: written by Torfs & Brauer, Hydrology and Quantitative Water Management Group, Wageningen University.
- Workshop on Data Visualization in R: by National Bioinformatics Infrastructure Sweden (NBIS), on behalf of SciLifeLab.
- R for Data Science (2nd Edition): written by Hadley Wickham, Mine Çetinkaya-Rundel, and Garrett Grolemund.
- Exploratory Data Analysis with R: written by Roger D. Peng, Johns Hopkins Bloomberg School of Public Health. 2020
- ggplot2: Elegant Graphics for Data Analysis (3e): written by Hadley Wickham, Danielle Navarro, and Thomas Lin Pedersen (2023).

17 May 2023